AlphaFold Outputs¶
Content of the output directory "per" query
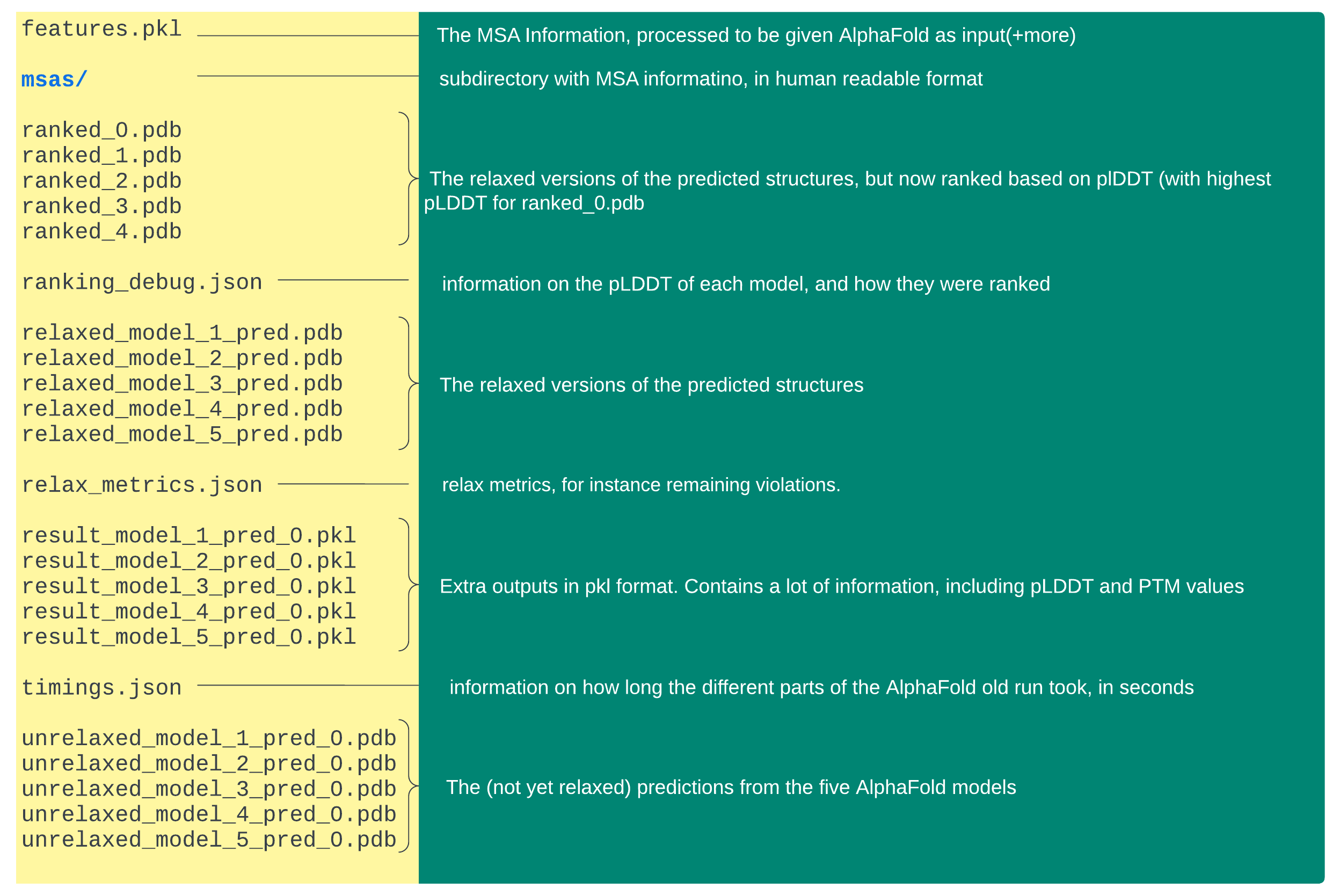
Each query will generate a subdirectory in output path with AlphaFold outputs. Below, a summary is given of their contents. The different file extensions are as follows:
.pdb– protein database format. These files contain the actual structures, including the pLDDT (local confidence) in the b-factor column.pkl– pickle data format, used in programming, but in a binary format an thus not readable from the command line.json– readable data format, often used in programming languages, but contents can be shown with cat and other commands msa files have .sto or .a3m file extensions, readable with cat and other commands
Note that by default, there are no image files attached in the output, for instance visualizing the pairwise PTM scores (global confidence).
Running thousands of queries ?. Keep track of Disk and Inode quota
-
The files that take up the most space are usually the MSA (depending on the actual MSA size, but this can take several GBs) and the
.pkldata files (grows quadratically with sequence length, with more than 1 GB reached for sequences of more than ~800 amino acids long) -
Use
nn_storage_quotato keep track of usage